Functional characterization of enhancer candidates in Drosophila embryos
We characterized 7793 transgenic Drosohila lines from the Vienna Tiles (VT) library, which is available from the Vienna Drosophila Resource Center (VDRC). Each line contains a transcriptional reporter construct with a ~2kb enhancer candidate DNA fragments (tiles), cloned in front of the minimal DSCP promoter and the yeast Gal4 gene as a reporter as in Pfeiffer et al. 2008. As all reporters were integrated into the same location of the fly genome, the VT library allows the assessment and direct comparison of enhancer activities across all candidates. All together these fragments cover approx. 13.5% of non-coding, non-repetitive D. melanogaster genome.
PCR verifications
The identity of 7478 lines (96%) was confirmed using PCR of genomic DNA (primers 5'-CGTATCACGAGGCCCTTTCGTCTTC-3' and 5'-GCACTTCGGTTTTTCTTTGGAGCACTT-3') isolated from transgenic flies followed by Sanger sequencing (the remaining 4% were either not tested or failed). In addition, for 182 randomly selected lines, we tested if the transgene is integrated at the correct position in the attP2 landing site using primers from Venken et al. 2006. As expected all 182 lines (100%) were integrated correctly suggesting that virtually all fragments reside in the attP2 landing site.
The pipeline for detection of enhancer activity
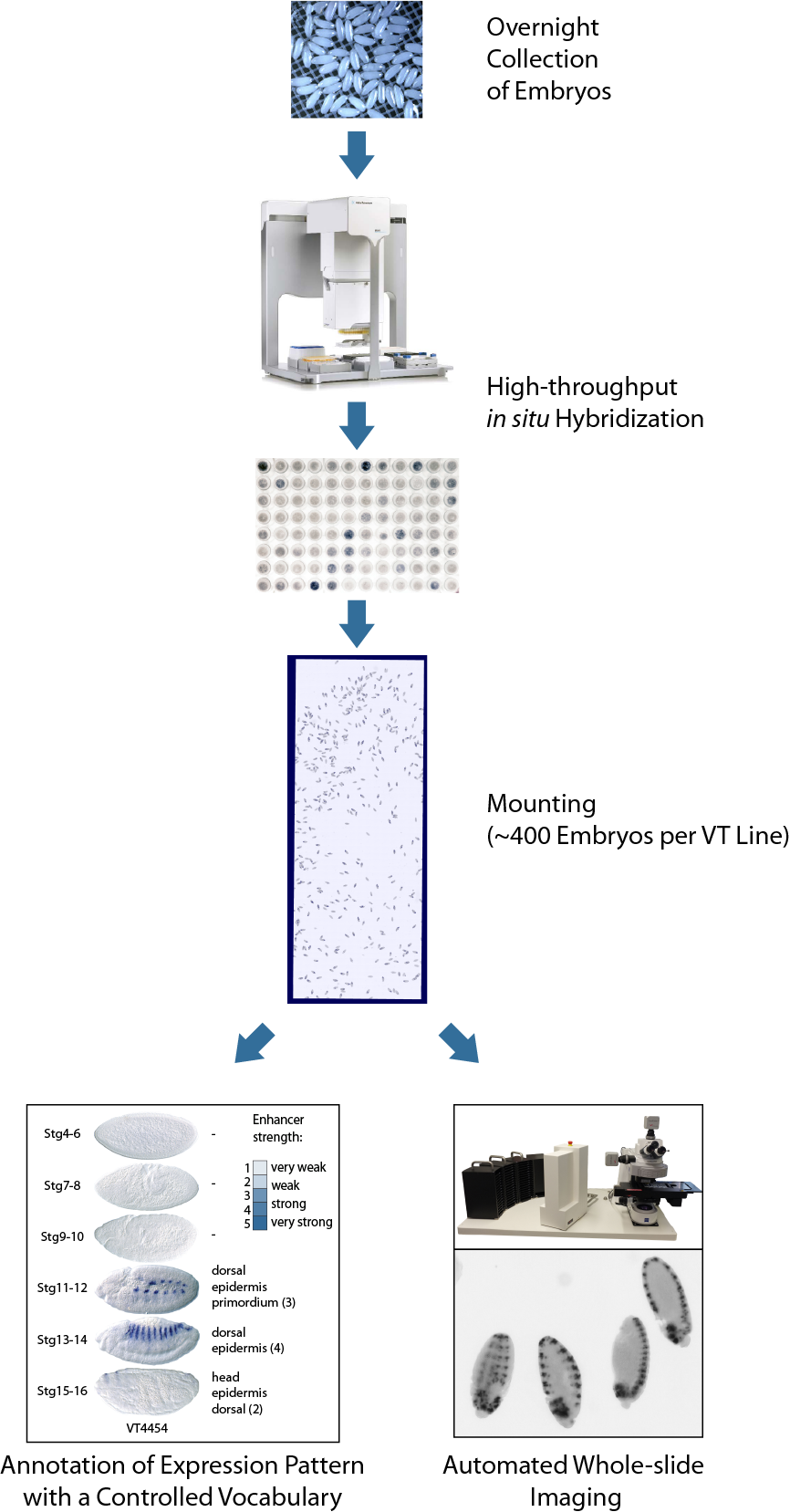
We developed a high-throughput pipeline, which allowed the robust and sensitive detection of enhancer activities in fly embryos. For each line we first harvested an overnight collection of embryos covering all stages of Drosophila embryogenesis. Approx. 400 transgenic embryos per each enhancer candidate were hybridized against a Gal4 antisense probe in 96-well format, mounted and imaged using automated whole-slide acquisition (see Figure).
Images
Whole slide images of stained transgenic embryos together with annotations are available at the pages of individual tiles, eg., VT013456. Be aware that except for stages 4-16, which we focus on, you can occasionally see embryos of earlier/later stages, and, that the stage 17 embryos often show staining artifacts. For slides that contained too few embryos of early stages (especially stages 4-6), we performed additional embryo collections, stainings, and imaged those slides separately.
Annotations
In addition to imaging, each line was manually inspected under the microscope and expression patterns in corresponding anatomical structures were annotated with a controlled vocabulary at 6 time intervals of embryonic development covering all major events of embryogenesis equivalent to the annotation of in situ gene expression patterns in BDGP. We use a few additional annotation terms where the original terms where too coarse — these are called original term subset.
Note that all coordinates refer to the dm3 assembly of the D. melanogaster genome. Please use the UCSC liftOver tool for coordinate conversion between different assembly versions.
More details can be found in E. Kvon et al. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature, 2014.
